Lesson 3: Tips for clear, reproducible code
Objectives:
- Understand the importance of documenting what is being done in your code
- Generate modular code
- Reduce repetition through the use of
loopsandfunctions
What is required for absolute reproducibility?
So far in this workshop we have discussed the set-up and use of RStudio projects, with a focus on how to generate various outputs (.pdf, .doc, .html) from an R Markdown document.
In addition to correctly formating our R Project, a completely reproducible analysis requires all of the information for somebody (you or otherwise) to re-do your analysis and get the same results. This includes:
- All raw data - saved in a file within the project directory
- All code used to clean and process raw data as well as generate any figures - scripts should be saved in a file within the project directory
- All code and software (specific versions and packages) for analysis - all package installation and loading steps should be outlined at the top of notebooks (even if not evaluated)
- There should be no external “by hand” steps in the analysis
The benefits of reproducibility are:
- You can work out what you did and understand why - decisions are explained
- Even if the (raw) data is updated, the same analyses can be completed
- You can pass your project on to others
- You can provide useful examples of your code to people who wish to inspect or continue your project
- When you are ready to publish it is easy to re-check the analysis
R Markdown files along with knitr can be used to neatly generate reports that follow each step of the analysis.
Style guidelines for neat coding
In addition to using R Projects and Markdown files to make our research reproducible, there are certain stylist aspects that make code easier to follow and read.
Why use a style guide?
- Clean code is easier to read and interpret - for you and others
- When code is clearer it is easier to spot mistakes
- Some style guide will help prevent potential problems e.g., avoiding
.in function names
There are several good style guides available, including Google’s R style guidelines, the Tidyverse style guide, Hadley Wickham’s R style guidelines, and the Bioconductor styel guide.
Some key points that are shared across guides:
- Use
<-not=for assignment - Guidelines for naming objects:
- Decide on a consistent naming style e.g., camelCase or snake_case
- Avoid spaces or dot (
.) and instead use an underscore (_) - Only use meaningful names - nouns for objects (e.g.,
todays_groups) and verbs for actions (e.g.,make_groups) - Avoid names of common functions e.g.,
mean
- Keep lines to 80 characters or less, as indicated by the margin column on your screen
- Put a space either side of binary operators (including
<-,+,-and=) - Do not put a space either side of a colon (
:)
Broader guidelines - omit needless code and avoid repetition. We will discuss how to do this in the next section of the workshop.
Reducing repetition by creating functions
You are likely already aware of and comfortable with functions in R - we’ve used several throughout this workshop. Functions allow us to automate common coding tasks in a more powerful way than copy-and-pasting. In addition to using functions that already exist in base-R or R packages, we can write our own functions.
The advantages of writing a function rather than repeating code:
- You can give a function a meaningful name to outline what the code is doing
- If you need to change something you will only need to update code in a single place
- Reduced chances of making a mistake e.g., copying a chunk of code without changing a variable name
When should you write a function?
It is good practice to write a function whenever we intend to run a set of commands more than twice.
Three steps to creating a function
There are three main things that we need to specify in order to generate a function:
- A meaningful function name
- A list of inputs to the function - the equivalent of standard arguments
- A set of commands to be packaged into the function body
function_name <- function(inputs) {function body}
Choosing a function name
The function name is what the function will be stored as within the R environment and how we will call the function when we wish to use it. As with all naming in R, function names should be clear, concise and meaningful. We usually use verbs in function names but nouns can also be used if they are descriptive and unambiguous. For example, if we want to create a function to calculate the circumference of a circle, it would be sensible to call this function circumference rather than function_1 or circumference_of_a_circle.
Defining the function inputs
The inputs to a function are the formal arguments, or ‘parameters’. These are the variables placed inside of the parentheses and separated by commas. When we call the function we will provide actual values to these arguments. In the example of circumference, the only input to our function will be the circle radius, r. This is the only variable in the equation C = 2 * pi * r. We will specify what to do with each input in the next section, the function body.
Writing commands in the function body
The function body is a set of commands provided inside of a pair of curly brackets ({}). These are the predefined set of commands that will be run every time we call our function.
## Define function
circumference <- function(r) {
2 * pi * r
}
## Use function
circumference(r = 2)## [1] 12.56637More complex considerations when creating a function
I want to set a default value for one of my inputs
To create a default value for one of the function inputs, simply include the value when defining inputs. The default values can still be over-written by specifying another value when calling the function. For example:
## Define function with default value of r
circumference <- function(r = 1) {
2 * pi * r
}
## Use function without specifying r
circumference()## [1] 6.283185## Use function and override default value of r
circumference(r = 5)## [1] 31.41593
I want my function to print value(s)
As you can see from the example above, as code is executed an output appears in the same way as when we execute code normally. Usually it is the last evaluated statement that will be returned. If we want earlier content to be returned we can use the explicit return function.
The return function is often combined with if or ifelse statements. For example, if an argument is missing we may wish to return a warning message to ourselves to remind ourselves that a default value is being used. Maybe in this case we would want to return an NaN value in response to a negative value of r, given that negative circumference is not possible.
circumference <- function(r = 1) {
2 * pi * r
if (r < 0) {
return(NaN)
}
}
circumference(r = -3)## [1] NaNI want my function to save value(s)
Sometimes we don’t just want our function to print the result but also save this in an object in our environment. There are several ways in which we can do this.
Let’s try using the normal assignment operator.
subtract_two_nums <- function(x, y) {
answer <- x - y
}
subtract_two_nums(x = 9, y = 6)The object answer only exists within the function and does not get saved within our R environment. This means that we can only use the answer object within the function. If we want to assign the answer to an object in the R environment we can use the assign function.
subtract_two_nums <- function(x, y) {
answer <- x - y
assign("result", answer, envir = .GlobalEnv)
}
subtract_two_nums(x = 9, y = 6)We are assigning the value of answer to an object called "result", which will be stored in the global environment. It is easy enough to print and assign the output of a function at the same time.
subtract_two_nums <- function(x, y) {
answer <- x - y
assign("result", answer, envir = .GlobalEnv)
print(answer)
}
subtract_two_nums(x = 9, y = 6)## [1] 3Challenge: Creating functions
Start by creating a simple function called add_seven which takes the argument x and both prints and saves the output of adding 7 to the value of x.
Solution
## Define function
add_three <- function(x) {
answer <- x + 7
assign("updated_x", answer, envir = .GlobalEnv)
print(answer)
}
## Test function
add_three(x = 8)## [1] 15Now try to create a function from scratch. Create a function to find the sum of all even integers between any two values.
Solution
cumulative_even_sum <- function(x, y) {
values <- x:y # first get all integers between the two values
even <- values[which(values %% 2 == 0)] # subset values that have a remainder of 0 when divided by 2
sum(even) # sum the even values
}
cumulative_even_sum(0, 10)## [1] 30Reducing repetitions by applying loops
Another way to reduce repetition in our code is through the use of loops. When you create a loop, R will execute all commands within the loop a specified number of times or until a condition is met. There are three main types of loop in R:
- the for loop
- the while loop
- the repeat loop
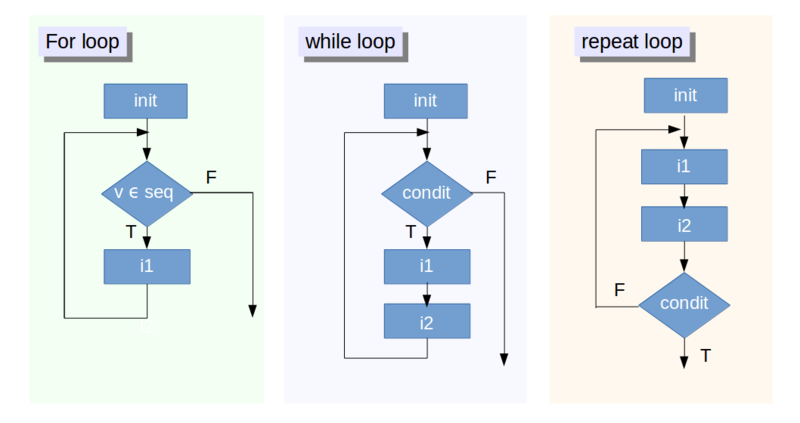
For loops - the most common R loop
A for loop is the most frequently used loop in the R language and is used to carry out a set of commands in an iterative manner over a collection of objects. This can be over each value of a vector, each column of a data frame, each component of a list etc. The loop will repeat the task a defined number of times.
for (variable in sequence) {expression}
The sequence is the collection of objects (eg., vector) over which the for-loop iterates. A variable is an item of that collection at each iteration, and the expression is a set of commands that we wish to apply to each variable.
Let’s look at a simple for loop.
for (x in 1:5) {
print(x)
}## [1] 1
## [1] 2
## [1] 3
## [1] 4
## [1] 5The expression inside of the loop, here print(x) is carried out iteratively for each value of x. In other words, the loop will first be completed using x = 1, then once it has finished another iteration begins with x = 2, then x = 3, until all variables in the sequence have been used. Even simple loops like this can be made useful in a real-world setting.
for (month in 1:5) {
print(paste("Month:", month))
}## [1] "Month: 1"
## [1] "Month: 2"
## [1] "Month: 3"
## [1] "Month: 4"
## [1] "Month: 5"for (month in 1:5) {
if (month < 3) {
print(paste("Winter"))
}
else {
print(paste("Spring"))
}
}## [1] "Winter"
## [1] "Winter"
## [1] "Spring"
## [1] "Spring"
## [1] "Spring"I want to store the output of my for loop
As was the case when we created our own functions, the results of a for loop are not automatically saved as an object in our environment. To do this we can append the results to an empty vector, which we define before the for loop.
month_vector <- c()
for (month in 1:5) {
month <- (paste("Month:", month))
month_vector <- c(month_vector, month)
}Challenge: storing loop outputs
Alter the seasons for loop to save a vector of length 5 which contains the outputs of our loop.
Solution
seasons_vector <- c()
for (month in 1:5) {
if (month < 3) {
seasons_vector <- c(seasons_vector, "Winter")
}
else {
seasons_vector <- c(seasons_vector, "Spring")
}
}I want to use a for loop on a list
Sometimes we want to do the same set of commands to different objects. For example, we may have a set of data frames that each hold the same experimental data taken from separate biological replicates. To do the same operations to these data frames, we could put these into a list and then loop over the lists.
Let’s create some data frames and store them in a list.
## Create example data frames
df_1 <- data.frame(month_vector, seasons_vector)
df_2 <- df_1
df_3 <- df_1
## Store data frames in a list
all_dfs <- list(df_1, df_2, df_3)
## Store new names
new_names <- c("updated_df1", "updated_df2", "updated_df3")A list can be used to store data frames, vectors, matrices - most objects. To loop over items in a list we can use indexing.
for (i in 1:length(all_dfs)) {
df <- all_dfs[[i]]
new_df <- df %>%
as_tibble %>%
mutate(weather = ifelse(seasons_vector == "Winter", "cold", "warm"))
assign((new_names[[i]]), new_df, .GlobalEnv)
}The lapply function
We can also loop through a list using the lapply function. The lapply function takes two arguments, X and FUN. The value of X is the sequence we want to apply to e.g., a vector or list. The FUN argument is short for function, this can be a pre-existing function or we can define a function here.
## Using lapply with a pre-existing function
lapply(X = c(1.34, 1.78, 2.34, 1.12), FUN = round)## [[1]]
## [1] 1
##
## [[2]]
## [1] 2
##
## [[3]]
## [1] 2
##
## [[4]]
## [1] 1## Using lapply with a user-defined function
lapply(X = c(1.34, 1.78, 2.34, 1.12), FUN = function(x){
x + 1
})## [[1]]
## [1] 2.34
##
## [[2]]
## [1] 2.78
##
## [[3]]
## [1] 3.34
##
## [[4]]
## [1] 2.12The output of lapply is a list - we can tell this from the double square bracket nomenclature. To do the same thing but have our output as a vector we can use sapply.
## Using lapply with a user-defined function
sapply(X = c(1.34, 1.78, 2.34, 1.12), FUN = function(x){
x + 1
})## [1] 2.34 2.78 3.34 2.12Challenge: Using loops and the apply family of functions Add another columns to each of the updated data frames to store numerical temperature values (make these up). Then use the apply functions (lapply or sapply) to loop over the updated data frames and output a vector containing the mean temperature.
Solution
## Put objects into a list
updated_dfs <- list(updated_df1, updated_df2, updated_df3)
new_names <- c("updated_df1", "updated_df2", "updated_df3")
## Create new temperature columns
for (i in 1:length(updated_dfs)) {
df <- updated_dfs[[i]]
df$temp <- c(4.8, 5.2, 13, 14.1, 12.7)
assign((new_names[[i]]), df, .GlobalEnv)
}
## Use sapply to get vector containing mean temperature
updated_dfs_v2 <- list(updated_df1, updated_df2, updated_df3)
sapply(X = updated_dfs_v2, FUN = function(x) {
x %>%
as_tibble() %>%
pull(temp) %>%
mean()
})## [1] 9.96 9.96 9.96